Viewing ENA Records¶
ENA Records¶
You can view any public ENA records using their Accession (unique identifier) or as the result of a search.
Exploring an ENA record¶
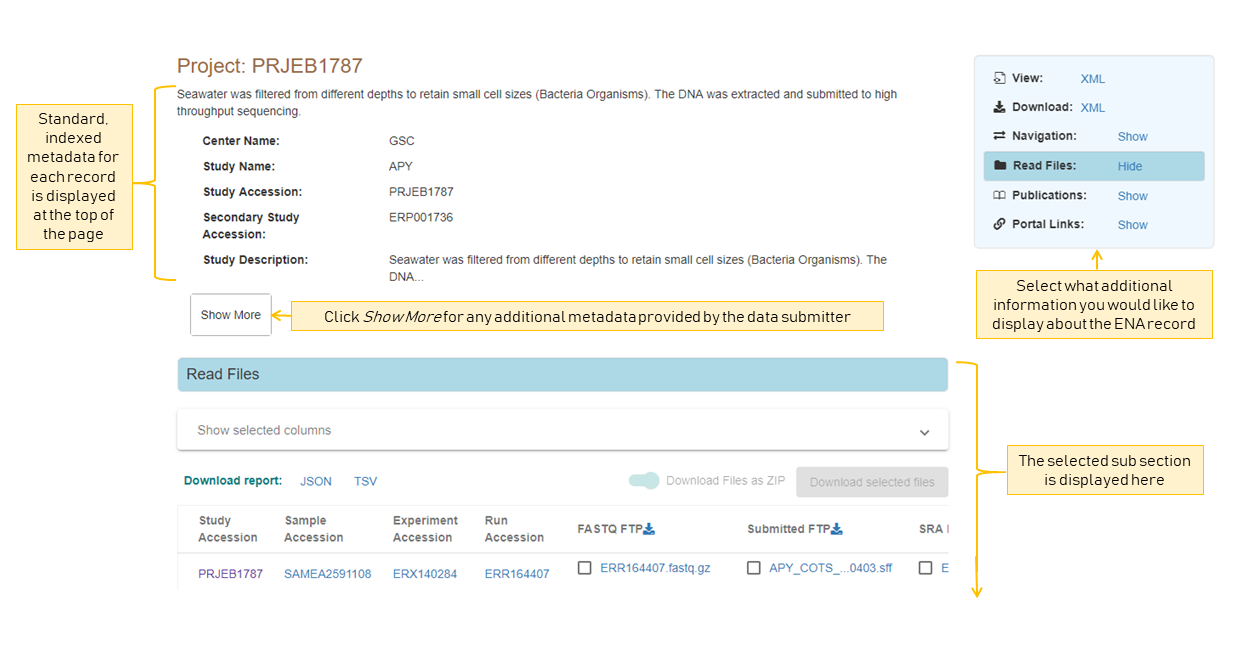
All records have a selection of standard, indexed metadata which tells you about the database entry. For example, where it is submitted, basic information on the record’s title and description etc. To show the full set of indexed metadata for this record, you can click Show More. For additional custom metadata provided by the submitter, you can view this in the ENA record XML.
To explore a record further, you can use the navigation box in the top right of the view to show/hide different additional subsections of information.
This gives you access to any other associated data, such as if a project has any data files or publications associated with it.
Any links that looks like accessions (a series of letters then numbers) will take you to an associated record.
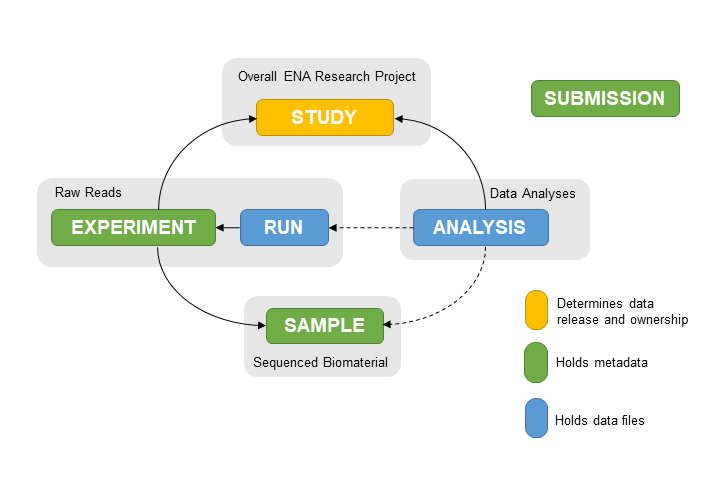
An ENA record can be one of the following types:
Record types¶
| Record Type | Description | Example accessions |
| Projects/Studies | Contains information on a biological
research project. This holds all the data
generated as part of this research.
|
PRJEB1787/ERP001736 |
| Samples | Represents biological samples collected
and sequenced in real life
|
SAMEA2620084/ERS488919 |
| Runs/Experiments | Hold raw read files and sequencing methods | ERR1701760/ERX1772048 |
| Analyses | Hold results files of analyses performed
on sequencing data and analysis methods
|
ERZ841272 |
| WGS contig set | Hold Whole Genome Sequencing contig sets
generated as part of a genome assembly.
|
|
| Assemblies | Represents an entire genome assembly and
holds any contig sets or sequence records
generated as part of the assembly
|
GCA_000001405.28 |
Assembled/Annotated
Sequences
|
Any sequence records from coding or
non-coding regions to full assembled
chromosomes
|
CM000667.2 |
| Taxon | The sequenced organism or metagenome of a
sample
|
Taxon:9606 |
| Sample Checklist | The checklist of metadata that the sample
was registered with
|
ERC000013 |
View and Download Links¶
This panel contains the different data formats available for the current record.
Contigset (WGS/TLS/TSA)¶
- View/Download flatfile for the master record describing the set
- Download the full set of sequences in one file (Using ftp:// is recommended for programmatic downloads)
- EMBL (from FTP) ftp://ftp.ebi.ac.uk/pub/databases/ena/wgs/public/cab/CABHOY01.dat.gz
- FASTA (from FTP) ftp://ftp.ebi.ac.uk/pub/databases/ena/wgs/public/cab/CABHOY01.fasta.gz
- FASTA (via API) https://www.ebi.ac.uk/ena/browser/api/fasta/CABHOY010000000
Project/Sample/Run/Analysis/Submission (XML types)¶
- View/Download metadata in XML format
Assembly (Genome Collection)¶
- View/Download metadata in XML format
- View/Download Sequence report as a text file
- Download all sequences in the assembly, including chromosomes, WGS set and scaffolds as one file
Read Files¶
Display and download any associated raw read files. Please refer to Archive Generated Files for more information about file formats.
There are several ways to download read files:
1. Using ENA File Downloader Command Line Tool¶
The ENA File Downloader is a new Java based command line application. You have to submit one or more comma separated accessions, or a file with accessions that you want to download data for. This tool allows downloading of read and analysis files, using FTP or Aspera. It has an easy to use interactive interface and can also create a script which can be run programmatically or integrated with pipelines.
Download the latest version from ENA Tools.
2. File Reports¶
You can download a Report of all the data displayed in the table or download files selected from the table. To download all files in the column, click the download icon in the table header.
To choose additional metadata to add to the table display and report, use the ‘Show selected columns’ expandable menu.
3. enaBrowserTools¶
You can also download files from ENA using the Python based scripts enaBrowserTools.
Analysis Files¶
Display and download any associated analysis files. There are three ways to download analysis files:
1. Using ENA File Downloader Command Line Tool¶
The ENA File Downloader is a new Java based command line application. You have to submit one or more comma separated accessions, or a file with accessions that you want to download data for. This tool allows downloading of read and analysis files, using FTP or Aspera. It has an easy to use interactive interface and can also create a script which can be run programatically or integrated with pipelines.
Download the latest version from ENA Tools.
2. File Reports¶
You can download a Report of all the data displayed in the table or download files selected from the table. To download all files in the column, click the download icon in the table header.
To choose additional metadata to add to the table display and report, use the ‘Show selected columns’ expandable menu.
3. enaBrowserTools¶
You can also download files from ENA using enaBrowserTools.
Publications¶
Explore publications that either cite the record or document the research where the record was generated.
This view provides links to the DOI or in some cases, a direct link to the PDF or article in Europe PMC.
Component Projects¶
In the case of an Umbrella Project (a project which is used to group many related sub-projects) there is the option to explore its Component Projects.
Component projects are the same as other project records in ENA but are grouped under one ‘umbrella’ meaning they will often have the same research motivation and will often represent a collaborative research effort.
Parent Projects¶
If a project has a parent project it is part of an Umbrella Project (a project which is used to group many related sub-projects).
Projects grouped under one ‘umbrella’ often have the same research motivation and will often represent a collaborative research effort. You can navigate to the parent project through this tab and view other related component projects through the ‘Component Projects’ tab.
Tax Tree¶
Here you can view the full tax tree of this taxon record.
From this view you can access all taxon records within this tax tree and explore ENA records that are registered with related taxa.
Click the arrows to expand the tree and explore the full lineage of the taxon.
Assembly Versions¶
If this assembly has been updated, you can view the past assembly versions here.
Assembly Statistics¶
Assembly statistics are generated for all assemblies submitted to INSDC.
Total Length (total sequence length) - total length of all top-level sequences.
Ungapped Length (total ungapped length) - total length of all top-level sequences ignoring gaps. Any stretch of 10 or more Ns in a sequence is treated like a gap.
Chromosomes & Plasmids (total number of chromosomes and plasmids) - total number of chromosomes, organelle genomes, and plasmids in the assembly.
Spanned Gaps - total number of gaps between contigs/scaffolds.
Unspanned Gaps - total number of unspanned gaps between scaffolds.
Regions/Patches/Alternative Loci - (number of regions with alternate loci or patches) - number of genomic regions that contain one or more alternate loci or patch scaffolds.
Scaffolds (number of scaffolds) - number of scaffolds including placed, unlocalized, unplaced, alternate loci and patch scaffolds.
Scaffold N50 - length such that scaffolds of this length or longer include half the bases of the assembly.
Contigs (number of contigs) - total number of sequence contigs in the assembly. Any stretch of 10 or more Ns in a sequence is treated as a gap between two contigs in a scaffold when counting contigs and calculating contig N50 & L50 values.
Contig N50 - length such that sequence contigs of this length or longer include half the bases of the assembly.
Chromosomes¶
When an assembly is is declared as assembled to full chromosome level on submission, chromosome sequences are generated for each chromosome submitted in the assembly.
These chromosomes are available as individual sequence records and can be explored in full here.
BlobToolKit¶
BlobToolKit is a set of computational tools developed to identify cross-species contamination within genome assemblies. A summary of results and graphics generated by BlobToolKit is displayed on the ENA browser to give data providers and consumers access to visualisation tools needed to identify contamination in public genome assembly data. BlobToolKit was developed by Richard Challis & Mark Blaxter at the University of Edinburgh.
For further information regarding BlobToolKit, please visit https://blobtoolkit.genomehubs.org.
Please send any questions or queries regarding BlobToolKit to blobtoolkit@genomehubs.org.
Checklist Fields¶
Sample Checklists are lists of fields that are required/recommended to be used during registration to describe samples (depending on the type of sample).
Explore the mandatory, recommended and optional fields defined as part of this checklist.
You can filter these fields further by requirement or by keywords in the field name or description.
In some cases, fields can be restricted by regular expression, a list of text choices, by valid taxonomy or by valid ontology terms.
3rd Party Curations¶
This tab presents the flow of 3rd party curations from the ELIXIR Contextual Data ClearingHouse (CDCH) data store.
The CDCH data store aims to provide a seamless method of exchange for curated contextual data available in external resources and community curation efforts, with ELIXIR data resources.